Scientists create a molecular toolbox for cracking protein postcodes
The team of Helle Ulrich at the Institute of Molecular Biology (IMB) in Mainz, Germany, has developed a new technique that allows scientists to tag a protein of interest with any one of three specific post-translational ubiquitin modifications. This technique, which they named “Ubiquiton”, opens a way for scientists to directly control the location and stability of cellular proteins, and to investigate how the cell’s own control mechanisms may go wrong in disease. The results of their study were published in the journal Molecular Cell.
For our cells to function smoothly, all of their proteins must be transported to the right locations, while proteins that are damaged or no longer needed must be removed. To achieve this, cells “earmark” their proteins with chemical tags called post-translational modifications, which act like postcodes stamped onto the protein. Some posttranslational modifications mark proteins for transport to specific locations in the cell, while others mark unnecessary proteins or those that have completed their tasks for degradation and removal.
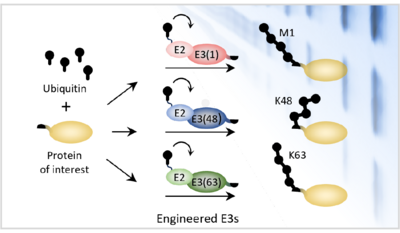
One important posttranslational modification is ubiquitin. Proteins can be tagged with ubiquitin in many different ways: they can be tagged with a single ubiquitin, or they can be polyubiquitylated by enzymes that join additional ubiquitin groups onto the first ubiquitin, like links in a chain. Depending on the attachment site at which the ubiquitin groups are joined together, the resulting polyubiquitin chain can be long and flexible, branched with multiple strands, or compact and folded. Scientists think that all of these different types of ubiquitin modifications act like distinct protein postcodes. However, this has been difficult to prove because scientists have had no way to directly manipulate polyubiquitin chains in the cell.
To address this need, Helle’s team have now developed a set of custom enzymes – in effect a molecular “toolbox” – that allows scientists to tag almost any protein with one of three specific polyubiquitin chains: a linear (M1) chain, a slightly kinked (K63-linked) chain, or a strongly kinked (K48-linked) chain. This toolbox, which they named “Ubiquiton”, was created by engineering a set of tailor-made enzymes using domains from human and yeast enzymes that are known to tag proteins with M1-, K63- or K48-polyubiquitin chains. To direct the custom enzymes towards specific target proteins, they utilised a pair of commonly-used protein domains that dimerise upon addition of the antibiotic rapamycin. By attaching one of these domains to the custom enzyme and one to the target protein, they were able to direct the custom enzymes to tag a target of choice simply by adding rapamycin to the growth medium. Finally, they employed another trick to coax the enzymes into starting a chain on the target protein.
Using the Ubiquiton system, Helle and her group showed that they could tag target proteins with each of the three polyubiquitylation chain types in vitro, as well as in yeast and even human cells. The system works for all kinds of proteins, both in the cytoplasm and the nucleus, and for membrane- and chromatin-associated proteins.
With this new system, Helle and her group could now clearly show that tagging proteins with K48-linked chains causes them to be degraded and removed from human and yeast cells, while tagging a plasma membrane protein with a K63-linked chain causes it to be taken up into the cell by endocytosis.
Helle says: “The Ubiquiton tool is inspired by the late physicist Richard Feynman, who stated: ‘What I cannot create, I do not understand’. We expect that our ability to build defined ubiquitin modifications on cellular proteins will help us to understand the mechanics of ubiquitin signalling in much more detail.” Christian Renz, who is the first author of the study, adds: “The system was born as a 'crazy idea' in a brain-storming session during a group retreat. Since then, it has come a long way and will hopefully become a valuable and widely-used research technology in the ubiquitin field.”
Through this new technique, Helle and her group have opened the way for researchers to deduce how cells direct their proteins to their correct destinations, as well as how this may be dysregulated in disease.
Further details
Further information can be found at www.sciencedirect.com/science/article/pii/S1097276523009619
Helle Ulrich is a Scientific Director at the Institute of Molecular Biology Mainz (IMB). Further information about research in the Ulrich lab can be found at www.imb.de/ulrich.
About the Institute of Molecular Biology gGmbH
The Institute of Molecular Biology gGmbH (IMB) is a centre of excellence in the life sciences that was established in 2011 on the campus of Johannes Gutenberg University Mainz (JGU). Research at IMB focuses on the cutting-edge fields of epigenetics, genome stability, ageing and RNA biology. The institute is a prime example of successful collaboration between a private foundation and government: The Boehringer Ingelheim Foundation has committed 154 million euros to be disbursed from 2009 until 2027 to cover the operating costs of research at IMB. The State of Rhineland-Palatinate has provided approximately 50 million euros for the construction of a state-of-the-art building and is giving a further 52 million in core funding from 2020 until 2027. For more information about IMB, please visit: www.imb.de.
Boehringer Ingelheim Foundation
The Boehringer Ingelheim Foundation is an independent, non-profit organization that is committed to the promotion of the medical, biological, chemical, and pharmaceutical sciences. It was established in 1977 by Hubertus Liebrecht (1931–1991), a member of the shareholder family of the Boehringer Ingelheim company. Through its funding programmes Plus 3, Exploration Grants and Rise up!, the Foundation supports excellent scientists during critical stages of their careers. It also endows the international Heinrich Wieland Prize, as well as awards for up-and-coming scientists in Germany. In addition, the Foundation funds institutional projects in Germany, such as the Institute of Molecular Biology (IMB) and the European Molecular Biology Laboratory (EMBL) in Heidelberg.
Press contact for further information
Dr Ralf Dahm, Director of Scientific Management
Institute of Molecular Biology gGmbH (IMB), Ackermannweg 4, 55128 Mainz, Germany
Phone: +49 (0) 6131 39 21455, Email: press(at)imb.de